The R package ecosystem is arguably one of the language’s strongest selling points. A package is typically a collection of functions, and sometimes data, along with documentation. R packages are community developed, typically open source, and serve to provide additional functionality or extend and build upon base R’s functionality.
The packages available range from huge projects used by millions of people such as dplyr and data.table, to much smaller ones catering to more niche interests. Use of some of the larger packages can have a big effect on the way a user writes and structures code, for example dplyr one of the tidyverse family of packages, encourages adoption of the authors ideals of tidy code and its use can influence the design of a project itself.
Other packages are less impactful and serve a more specific purpose. If you want to access open data from NHS Scotland for example, you could read the API documentation, learn how to use the httr package, and write bespoke code to import the data. Alternatively, you could install odns and have it all done for you (disclaimer: I wrote odns).
Using packages is a fundamental part of programming with R for many users, but for those just starting out its important to understand what they’re all about.
R packages
The official documentations states that ;
Packages provide a mechanism for loading optional code, data and documentation as needed. The R distribution itself includes about 30 packages.
At a basic level we can simply think of packages as a collection of functions and data with documentation. Typically users install a package in order to make use of functionality that isn’t provided by base R and to avoid having to write their own solutions to common and/or complex problems.
Anybody can write a package and use it locally, you can also share a package you wrote with friends and colleagues. Some may go on to make the packages they write available publicly on services such as GitHub, and those looking to reach a wider audience may decide to publish their package to CRAN or Bioconductor.
Regardless of how widely a package is intended to be used, R actually enforces quite a strict structure for packages in comparison to other programming languages. The benefit of this is that packages are largely easy to use and understand, and as you gain experience, easier to dissect and explore the source code.
Package sources
CRAN
The Comprehensive R Archive Network (CRAN) is the official repository. It hosts a large number of packages, has strict criteria around submission, and packages published there are thoroughly tested and must undergo a manual review process.
When using the install.packages() function, the default settings look for the package on CRAN.
Bioconductor
Biocondutor hosts software tailored to performing reproducible analysis of data with a biological focus, including the analysis of DNA microarray, sequence, flow, SNP, and other data. Like CRAN it also has a review process for submissions.
GitHub
Whilst other hosting services are available, GitHub is one of the most popular for R development. Anybody can host code on GitHub and there is no review or submission process. GitHub hosts development versions of many packages that are also on CRAN or Bioconductor.
Locally (zip or tar.gz file)
If a package has been written or developed locally it may be shared with you in a zip or tar.gz archive.
Installing packages
Install from CRAN
Installing packages from CRAN is pretty straightforwards. You pass a string specifying the package name as the pkgs argument of install.packages(), and if the package exists on CRAN it will be downloaded and installed along with other R packages that the target package depends on, imports, or links to (using the default options).
To install the odns package;
install.packages("odns")It’s also possible to install multiple packages at once.
install.packages(c("odns", "httr"))CRAN has mirrors all over the world, near identical copies of the repository hosted across multiple servers. Following a fresh install, using install.packages() may return a prompt asking that you select a mirror. Most people should simply select option ‘1’ (“https://cloud.r-project.org”), which defaults to use a mirror near to you.
You can also set the mirror of choice at any time the chooseCRAN() function.
--- Please select a CRAN mirror for use in this session ---
Secure CRAN mirrors
1: 0-Cloud [https]
2: Australia (Canberra) [https]
3: Australia (Melbourne 1) [https]
4: Australia (Melbourne 2) [https]
5: Australia (Perth) [https]
...
Selection: 1RStudio users also have the option to install packages through the user interface. Clicking Tools > Install Packages... opens up the relevant window, where the package name can be entered, before clicking Install.
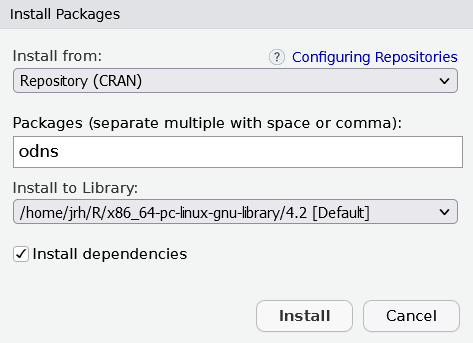
Install from Bioconductor
Installing from Bioconductor requires an additional package BiocManager, which can be installed from CRAN. Once BiocManager is installed, you can install Bioconductor packages using BiocManager::install().
install.packages("BiocManager")
BiocManager::install("GenomicFeatures")Install from local archive
If you have an R package as an archive (zip or tar.gz), these can be installed using the install.packages() function with a few additional options specified.
install.packages("path_to_file/odns.tar.gz", repos = NULL, type = "source")Install from GitHub
There are a couple of options for installing packages directly from GitHub, but the simplest is to use the remotes package, which can be installed from CRAN. Once remotes is installed, you can install packages from GitHub using remotes::install_github().
install.packages("remotes")
remotes::install_github("jrh-dev/odns")
Warning: Using any R package, whatever the source, involves an element of trust. Any package you install could potentially contain malicious code, but there are steps which can be taken to mitigate the dangers.
CRAN packages must undergo a series of checks upon submission, new submissions are carefully scrutinised, and packages on CRAN are subjected to ongoing testing on multiple platforms. The CRAN submission process does not provide a watertight guarantee of safety, but it does present a serious obstacle for potential bad actors. Bioconductor's own submission process presents a similar deterrent to CRAN.
Installing directly from GitHub or other repositories is inherently risky. There are no checks or tests performed on code hosted on GitHub outside of those chosen by the author. Whilst malware is against GitHubs terms of service, it takes time and reports for it to be identified. Installing popular packages from GitHub is not without risk either. Common approaches to spreading malware include attempting to direct users to install packages from apparently legitimate links such as tidiverse/dplyr.
In the past, developers credentials have been compromised and legitimate accounts under the control of bad actors used to push malicious code. In recent years a small number of developers have turned rogue and intentionally poisoned their own repositories. Thankfully, none of these high profile incidents have affected R directly.
Malicious additions and changes to popular open source projects should be caught quickly, but whilst we would expect them to be caught before they reach CRAN, they may already have been installed from GitHub many times.
GitHub is a fantastic resource and users should not be discouraged from using it, or from testing development versions of packages. However, installing packages from GitHub should be considered an option primarily for intermediate to advanced users with the skill to understand the source code within the packages being used.
Personally, I avoid using development or non-CRAN/BioConductor versions of packages in anything other than a sandbox environment. If there were no alternative packages available and I could not write the functionality myself, I would prefer to fork the repository, undertake a full code review, and then use the forked version in my own project.
Using packages
Once a package is installed you can start using it right away. Without loading the package, you can access its functions with ::.
odns::all_packages()
#' package_name package_id
#' covid-19-vaccination-in-scotland 6dbdd466-…
#' enhanced-surveillance-of-covid-19-in-scotland 3c5231ee-…
#' hospital-onset-covid-19-cases-in-scotland d67b13ef-…
#' weekly-covid-19-statistical-data-in-scotland 524b42b4-…
#' covid-19-in-scotland b318bddf-…
#' … with 85 more rowsTrying to call the function by name, without first loading the package returns an error.
all_packages()
#' Error in all_packages() : could not find function "all_packages"
#' To load the package and access it’s functions without the ::, you can use the library() function. The package name is passed without quotes as the package argument.
library(odns)
all_packages()
#' package_name package_id
#' covid-19-vaccination-in-scotland 6dbdd466-…
#' enhanced-surveillance-of-covid-19-in-scotland 3c5231ee-…
#' hospital-onset-covid-19-cases-in-scotland d67b13ef-…
#' weekly-covid-19-statistical-data-in-scotland 524b42b4-…
#' covid-19-in-scotland b318bddf-…
#' … with 85 more rowsWhen to use :: and library() is a matter of personal preference. Loading a large library with many functions to use one specific function is often not necessary, especially if the functions within that package mask some others that you do want to use. Google’s R style guide provides a useful take on the issue.
Qualifying namespaces
Users should explicitly qualify namespaces for all external functions.
# Good purrr::map()We discourage using the @import Roxygen tag to bring in all functions into a NAMESPACE. Google has a very big R codebase, and importing all functions creates too much risk for name collisions.
While there is a small performance penalty for using ::, it makes it easier to understand dependencies in your code. There are some exceptions to this rule.
When importing functions, place the @importFrom tag in the Roxygen header above the function where the external dependency is used.
- Infix functions (%name%) always need to be imported.
- Certain rlang pronouns, notably .data, need to be imported.
- Functions from default R packages, including datasets, utils, grDevices, graphics, stats and methods. If needed, you can @import the full package.
Updating packages
Over time new versions of packages are released. RStudio users can navigate to the ‘Packages’ pane, and click on the ‘Update’ button to perform a check for newer version of currently installed packages. This allows you to select any packages that you then want to update.
The old.packages() function provides a programmatic way to identify installed packages that have newer versions available.
old.packages()
#' Package LibPath Installed Built ReposVer
#' MASS "MASS" "/usr/lib/R/library" "7.3-58" "4.2.1" "7.3-58.1"
#' Repository
#' MASS "https://cloud.r-project.org/src/contrib"Having identified packages that can be updated, using install.packages() will install the latest version.
install.packages("MASS")Caution: When a package is updated it may fix bugs, add new functionality, or offer performance improvements. However, it may also change how some functions work, or remove them entirely. If you update a package that is used in a repeated workflow then this may lead to bugs, or your code may stop working altogether. Always test your workflows when updating packages.
When to use a package
The wide range of high quality packages available for R are one of its many strengths and a key element of its continued success. However, it is also important to consider dependencies and reproducibility. Adding packages that aren’t required, or that have easy to use alternatives in base R increases the amount of maintenance required for your code base over time. It also makes it harder for others to run your code.
Try to use packages sparingly and look critically at whether a package provides the best solution to a problem.
Viewing documentation
RStudio users can access the documentation for any installed package with ??.
??odns| Search Results | Package Vignette |
|---|---|
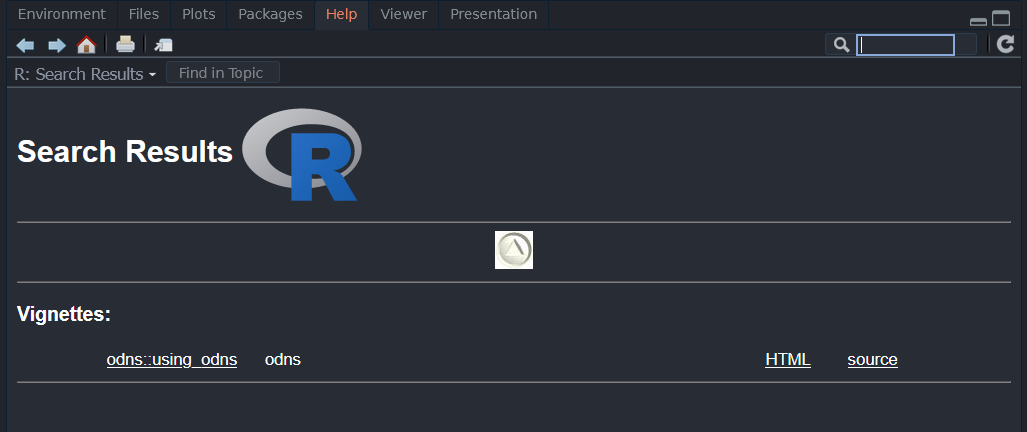 |
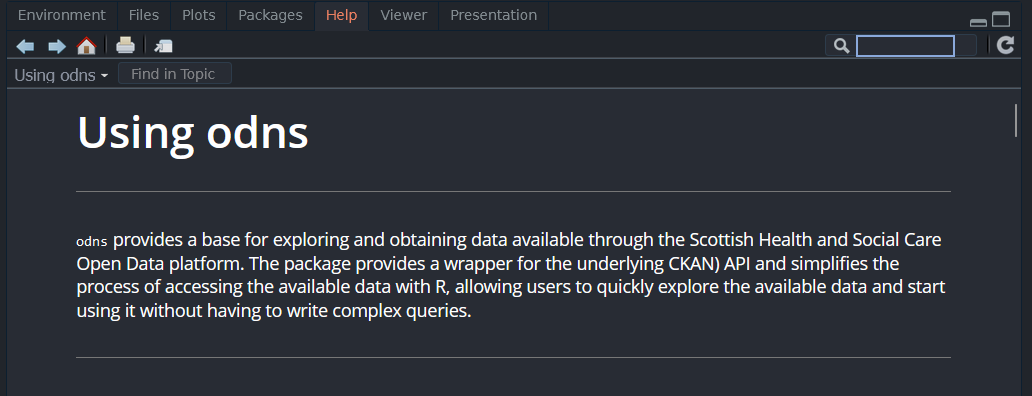 |
The documentation for functions or data within a package can be obtained in the same way.
??odns::all_packages
Next steps
Once you’re comfortable with installing R packages, it’s time to start exploring. Worthy mentions include;
-
The tidyverse, a collection of packages in reality, highly opinionated, but very popular.
-
data.table offers unparalleled performance and a style more in line with base R.
-
lubridate provides useful functions for dealing with dates.
-
ggplot2, the essential plotting package for R.